[ad_1]
The PLCO trial
Study participants were from the NCI PLCO Cancer Screening Trial, a large, randomized trial designed to evaluate if screening for prostate, lung, colorectal, and ovarian cancers lead to mortality reduction for these diseases9,10,11. Almost 155,000 men and women aged between 55 and 74 years were enrolled from 1993 to 2001 at 10 screening centers across the United States (Birmingham, Alabama; Denver, Colorado; Detroit, Michigan; Honolulu, Hawaii; Marshfield, Wisconsin; Minneapolis, Minnesota; Pittsburgh, Pennsylvania; Salt Lake City, Utah; St. Louis, Missouri; Washington, DC). Approximately half of the participants were randomized to the intervention arm and underwent cancer screening, while the other half were in the control arm and received standard medical care. Several self-administered questionnaires were administered at baseline and during follow-up, which collected information on demographics, medical history, family history and various lifestyle and dietary risk factors. Information from these questionnaires have been aggregated and harmonized to produce traits and covariates used in the PLCO Atlas genetic association tests. Blood was collected from screening-arm participants at baseline and at each annual screening visit for up to 5 additional years. In addition, buccal cells were collected from 2000–2003 from control arm participants and again in 2018 from participants in both arms. Cancer incidence and mortality outcomes have been tracked longitudinally with a median follow-up length >18 years for cancer incidence (approximately 44,000 cancers through 2017) and >19 years for deaths (approximately 57,000 deaths through 2018). All cancer diagnoses were confirmed by medical record review and/or via linkage to cancer registries, as previously described12,13. All participants provided written informed consent and the study was approved by the Institutional Review Boards at the National Cancer Institute and the 10 screening centers. Additional information about the cohort can be found at https://cdas.cancer.gov/learn/plco/home/.
GWAS data
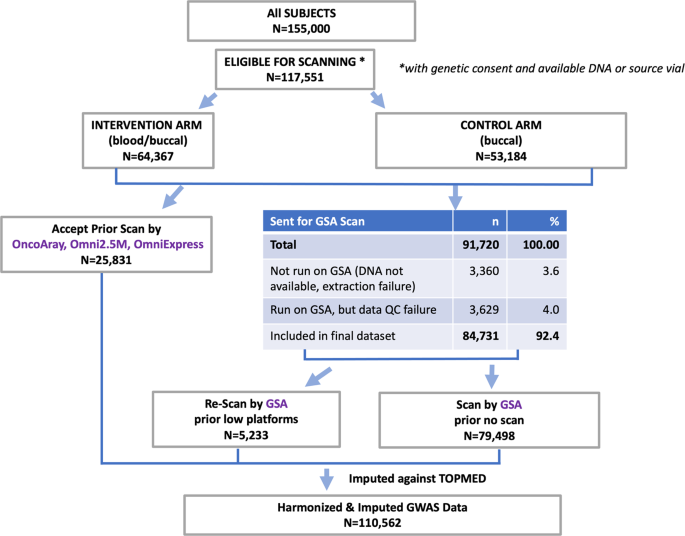
The PLCO Atlas genotyping project sought to genotype all PLCO participants with genetic consent and available DNA or source vial (N = 117,551) (Fig. 1). These participants were from the screening arm (N = 64,367) with blood and buccal source material and the control arm (N = 53,184) with only buccal source material. The Atlas project combined genotyping data previously generated by high density arrays for 25,831 participants (OncoArray, Omni2.5 M, and OmniExpress) as part of prior GWAS scans1,2,3,4,5,6,7,8 with a new round of genotyping using the Illumina Global Screening Array (GSA) for 84,731 participants who had low-density genotype data (n = 5,233) or no prior genotyping (n = 79,498).
Samples from a total of 91,720 participants were processed for GSA genotyping. DNA extraction was performed using appropriate chemistry based on source material type and automated on the KingFisher Flex Purification System. Extraction protocols were followed using standard operating procedures developed internally in the NCI Division of Cancer Epidemiology and Genetics Cancer Genomics Research (CGR) Laboratory. The predominant DNA sample source was buccal cells (48.4%), followed by buffy coat (39.8%), whole blood (4.2%), and buffy coat and red blood cells (1.4%), as well as previously extracted DNA (2.6%). Of the 91,720 participants whose samples were processed, 3,360 (3.6%) individuals were not genotyped on GSA due to insufficient DNA extracted (N = 2,313) or insufficient material from previously extracted DNA (N = 1,047). In addition, a total of 3,629 (4.0%) individuals were excluded from the final dataset due to quality control failures described below and summarized in Fig. 2a,b, resulting in a total of 84,731 PLCO participants successfully genotyped by GSA.
GSA genotyping was performed at the NCI Division of Cancer Epidemiology and Genetics CGR Laboratory according to Illumina protocols and following internal standard operating procedures. The CGR has extensive experience performing high-throughput Illumina bead-based genotyping having previously genotyped hundreds of thousands of samples. Initial GSA genotyping resulted in an overall failure rate of 1.5% for blood-derived DNA and a failure rate of 13% for buccal-derived DNA. After additional DNA extraction and genotyping to recover sample failures, genotyping was fully completed for a total of 84,731 GSA genotyped individuals.
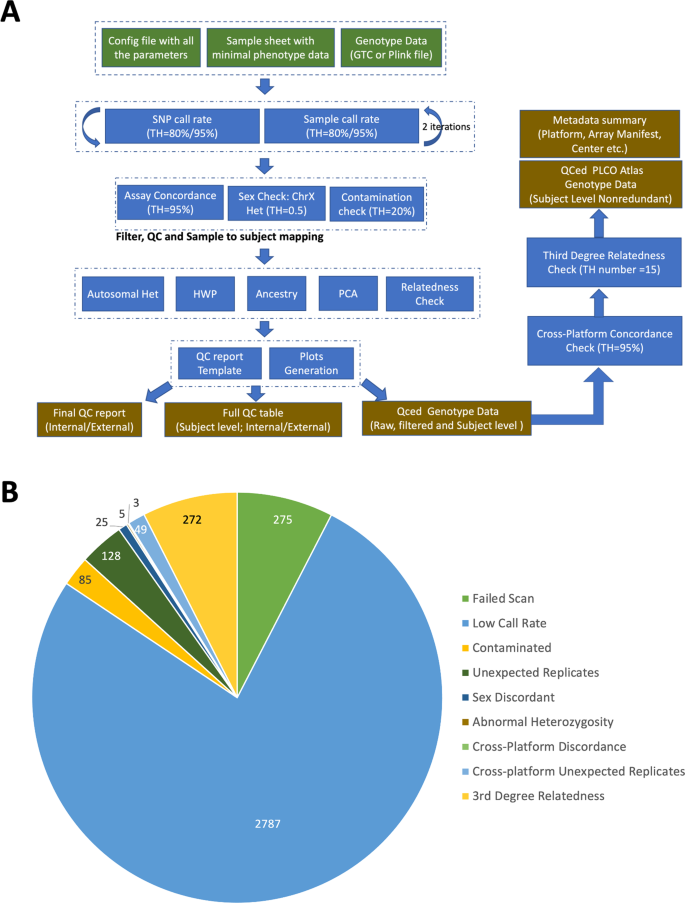
Extensive quality control filtering was performed for each array to ensure a set of high-quality genotype data for subsequent imputation and association analyses. Detailed quality control steps and the reasons and numbers of exclusions for the GSA platform are described below and summarized in Fig. 2a,b, respectively. For subjects genotyped on GSA, 275 subjects failed to produce valid output files (either .idat and/or .gtc files) during array processing and were excluded from the study. Next, 2,787 subjects were removed after applying a two-stage filter by a completion rate threshold of 0.8 for samples and 0.8 for loci, followed by a further 0.95 filter for samples and 0.95 filter for loci. A sample contamination check was performed using VerifyIDintensity, in which 85 subjects with greater than 20% estimated contamination were removed. Pairwise genotype concordance for all subjects was assessed to identify unexpected replicates, where subjects with a genotype concordance greater than 95% for a set of LD-pruned SNPs were considered replicates. After reviewing concordance check results against the enrolled phenotype data, a total of 128 subjects were removed. Sex was verified by comparing the reported sex with the observed sex based on X chromosome method-of-moments F coefficient from PLINK. The F coefficient is expected to be close to 0.0 for males and 1.0 for females with our threshold set to 0.5 for separating the two populations. Samples that failed the sex concordance check were subject to additional screening for sex chromosome aneuploidies by STR profiling using the AmpFLSTR Identifier assay, resulting in a total of 25 subjects excluded due to sex discordance. Further, a total of 5 subjects were identified to have abnormal heterozygosity by using absolute values from PLINK method-of-moments F coefficients greater than 0.2. Pairwise genotype concordance for all subjects from different datasets within the same platform and across different platforms was also assessed to identify cross-dataset and cross-platform discordant expected duplicates (n = 3) and unexpected replicates (n = 49). Additionally, relatedness was examined using Plink IBS/IBD tests. A total of 272 subjects from the GSA platform were identified with genetic relationships at the pi_hat threshold (0.1) and were removed. Consequently, a total of 3,629 individuals were excluded from the GSA dataset due to quality control failures (Fig. 2b).
Cumulatively, samples from all platforms were filtered to remove abnormal levels of heterozygosity (N = 12), sex discordance (N = 31), within-dataset unexpected duplicates (N = 130), discordant expected duplicates (N = 14), cross-dataset and cross-platform unexpected duplicates (N = 126), and relatedness check (N = 291). A total of 47 subjects with sex-chromosome abnormalities were retained in the dataset for downstream imputation.
After applying QC exclusions to each array, a total of 112,065 DNA samples genotyped across 110,562 unique individuals on a modern, high-density Illumina genotyping array remained (Table 1). For participants genotyped on multiple genotyping arrays (N = 1,192), only genotype data from one array was included in the Atlas project following the prioritization of Global Screening Array (GSA) > OncoArray > Omni2.5 M > OmniExpress (OmniX) to ensure non-redundant subject-level genotyping data. The predominant genotyping array was the GSA (N = 84,731), followed by the OncoArray (N = 16,893), Omni2.5 M (N = 7,211) and OmniX (N = 1,727).
Genetic ancestry for PLCO Atlas participants was determined using GRAF (https://github.com/ncbi/graf) on a set of 10,000 pre-selected fingerprinting variants. GRAF assigned individuals into the following 9 ancestral groups: “African”, “African American”, “East Asian”, “European”, “Hispanic1”, “Hispanic2”, “Other”, “Other Asian”, and “South Asian”. Hispanic1 included individuals of Dominican or Puerto Rican ancestry whereas Hispanic2 included individuals of Mexican or Latin American ancestry. For parsimony and to facilitate downstream analyses, we merged “African” and “African American” into a “African American (Combined)” group and also “East Asian” and “Other Asian” into a “East Asian (Combined)” group. The largest ancestral sets in the PLCO Atlas included European (N = 100,448), African American (Combined) (N = 4,576) and East Asian (Combined) (N = 3,528).
For genotype imputation, we used the TopMed reference panel on the Michigan Imputation Server, which is accessible on the Michigan Imputation Server to all TopMed collaborators. To prepare for genotype imputation on the Michigan Imputation Server (MIS, https://imputationserver.sph.umich.edu), we filtered variants with minor allele frequency ≤ 0.01, variant-level missingness ≥ 0.05, and Hardy Weinberg equilibrium exact test p-value ≤ 1 × 10−6 from the imputation input. Data from each genotyping platform were then analyzed using a community-recommended script for aligning data to reference datasets (HRC-1000G-check-bim.pl, from https://www.well.ox.ac.uk/~wrayner/tools/). The script was modified to support TOPMed 5b as a reference panel using a pre-existing test imputation with 1000 Genomes Project subjects versus the TOPMed 5b reference panel. Data were uploaded to the MIS in GRCh37/hg19 and lifted over by the MIS. Pre-phasing using phased reference data from TOPMed release 5b was conducted using EAGLE 2.4. Imputation was conducted against the same reference panel using minimac4 (https://genome.sph.umich.edu/wiki/Minimac4). The “Population” option was set to “EUR” for GSA batches 1–4 that included European ancestry samples, while the option was set to “Other/Mixed” for all other imputations, which consisted of non-European samples or samples of uncharacterized ancestries. The PLCO imputation process took place over several months and was run in different rounds over the span of those months. In total, 110,562 subjects were successfully imputed to the TOPMed 5b reference panel.
Following MIS imputation, raw imputation data were partitioned into subsets according to predicted GRAF genetic ancestry groups to estimate ancestry-specific imputation quality. Ancestry and chip combinations with less than 100 individuals were deemed to have insufficient sample sizes for association testing and removed. After partitioning by ancestry and recomputing imputation quality Rsq values, each platform and ancestry pair was cleaned according to the filtering method described by Kowalski et al.14. Briefly, all variants with Rsq <0.3 were removed to be consistent with traditional quality filters. Remaining variants were then partitioned into minor allele frequency (MAF) bins (<0.05%, 0.05–0.2%, 0.2–0.5%, 0.5–1%, 1–3%, 3–5%, and >5%) and each bin was filtered, starting at the variant with the lowest Rsq, until the average Rsq of remaining variants within the corresponding MAF bin was at least 0.9. In total, more that 78,000,000 high-quality imputed variants were available for association testing. In addition, we observed high concordance between high quality imputed SNPs from the GSA with genotyped variants present on the OmniExpress arrays, with a median correlation of 1.00 and a mean correlation of 0.984.
Filtered imputed data by platform and ancestry were then converted to bgen format (v1.2) for compatibility with BOLT-LMM and SAIGE for association testing. The resulting final imputed PLCO Atlas Project dataset for association analyses is detailed in Table 2.
Association analysis
Association analyses on the autosomes and X chromosome were carried out using the PLCO pipeline hosted on GitHub (https://github.com/NCI-CGR/plco-analysis). All variants in non-PAR regions of the X chromosome in males were handled by coding these variants as 0/2. Quantitative phenotypes with a sample size of at least 3,000 subjects were analyzed by BOLT-LMM v2.3.415, using linear mixed models on variants with MAF >0.01. The top 20 principal components (generated separately by ancestry) were included as adjustment variables, as well as participant’s age, sex, and study center. Healthy subjects free of any cancer diagnoses throughout the follow-up period were treated as controls for all cancer analyses. Binary and categorical phenotypes were analyzed with SAIGE 0.43.216. We required more than 1,000 subjects and at least 50 cases for each SAIGE phenotype tested. At the variant level, a minimum variant count of 5 and a MAF >0.01 were required for testing. Association analyses were run separately for every GRAF-defined ancestry group, genotyping array, and imputation group. Ancestry-specific results were aggregated by meta-analysis to create overall summary results as well as sex-specific summary result files. Quantile-quantile (Q-Q) plots were generated and lambda values were calculated for each phenotype by linkage disequilibrium score (LDSC) regression17.
After association analyses using BOLT-LMM or SAIGE, the SNP column of the GWAS summary files were annotated by a custom tool (https://github.com/NCI-CGR/annotate_rsids_from_linker.git), in the format of rsid:otherAllele:testedAllele (or chr:pos:otherAllele:testedAllele if there was no matching rsid). Population-specific data from the 1000 Genomes Project imputed with the TopMED imputation 5b panel was used to annotate allele frequencies for each tested variant in the GWAS summary statistics using the annotate_frequency program (https://github.com/NCI-CGR/annotate_frequency). Association analyses for every GRAF-defined ancestry group were run separately for each genotyping array and imputation group. While the genotyping arrays and imputation procedures we implemented in the PLCO Atlas captures trait and disease associations with common variants shared across ancestries, associations with population-specific variants and variants with ancestry-specific differences in allele frequencies may not be well captured by this approach.
Currently the PLCO Atlas project hosts association results for 90 diseases and traits, including a comprehensive list of cancer types and subtypes defined by organ site, etiology, and pathology (Table 3). For example, in addition to overall female breast cancer, we’ve included invasive, in situ, ductal, lobular, tubular, ER positive, ER negative, PR positive, PR negative, ER positive or PR positive, ER negative and PR negative, HER2 positive, HER2 negative, ER, PR, and HER2 triple-negative, Grade III or Grade IV, Grade II, and Grade I breast cancer. By etiology, we’ve performed GWAS analyses for smoking-related, alcohol-related, obesity-related, height-related, physical activity-related, diabetes-related, and infection-related cancers (overall, and by HPV- or H. pylori-). For smoking-related cancers, for example, we’ve considered cancers of bladder, ureter, kidney, lip, oral cavity, oropharynx, nasopharynx, hypopharynx, larynx, nasal cavity, paranasal sinuses, colorectum, esophagus, gastric, liver (excluding intrahepatic bile duct cancer), lung, myeloid leukemia, ovarian (mucinous), pancreas, and uterine cervix. By pathology, we’ve organized cancers into solid tumors (e.g., carcinomas, sarcomas, or urothelial cancers) and hematologic cancers (e.g., lymphoid or myeloid). Within carcinomas, we further broke down to adenocarcinomas (excluding mixed adenocarcinoma), endocrine or neuroendocrine, and squamous cell cancers.
We also include GWAS association results for key cancer risk factors such as baseline status of body mass index, height, cigarette smoking for ≥6 months (never, ever), and cigarette smoking categories (never, former, current), caffeine consumption from diet, and male pattern baldness at age 45, as well as baseline measures of serum PSA level and serum CA-125 level. These initial traits were selected based on available previous data and represent binary, categorical, and continuous traits for the purpose of analytical pipeline development and validity checking. Analyses of additional traits are in progress and association results will be publicly posted as they become available.
Summary statistics
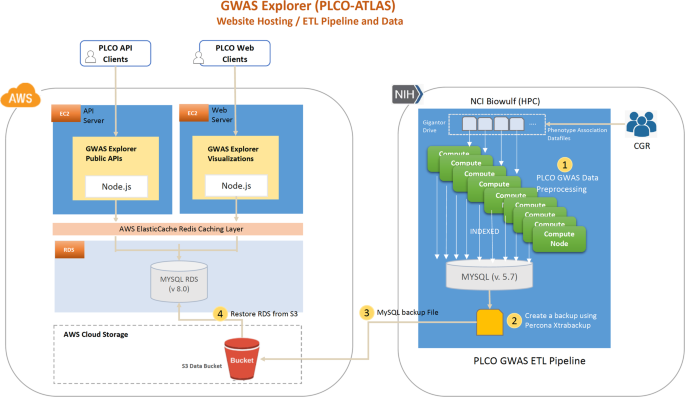
After association testing and annotation, summary statistic data was imported into a primary MySQL instance using an import script run on the National Institutes of Health (NIH) High Performing Computation Biowulf cluster (https://hpc.nih.gov/) that imported and aggregated participant phenotype metadata and variant association data. Using several parallel processes, each phenotype’s variant association data was aggregated and then indexed. Specific plot views for data visualization, such as the single chromosome summary view in the Manhattan plots and the q-q plots, were generated in this import and indexing process. The results were then pooled into the primary MySQL instance where a snapshot was created in Biowulf using Percona Xtrabackup tool. The snapshot was then uploaded to an Amazon Web Services (AWS) Simple Storage Service (S3) cloud bucket where it was restored to AWS’s Relational Database Service (RDS).
All PLCO Atlas summary statistic data is publicly posted on the GWAS Explorer (Fig. 3). The GWAS Explorer is hosted on AWS. It consists of two AWS EC2 servers, an AWS RDS instance, an AWS ElastiCache instance, and an NCI on-premises download server. The website and API are served by each of their own dedicated AWS EC2s. All PLCO data is hosted in a single AWS RDS MySQL instance, which can be scaled-up or duplicated if needed. The GWAS Explorer backend is hosted by Fastify NodeJS, a web application framework like the popular Express framework but optimized for faster API performance. All API routes and database queries defined and utilize MySQL database query logic. Website (internal) requests are routed to a dedicated web server and public API requests are routed to a separate dedicated API server to reduce load on the webserver during periods of high usage. Public API routes are documented with Swagger UI (see Data Records). Download requests are routed to a dedicated local NCI download server to reduce egress costs. Additionally, a cache layer is configured using Redis and AWS ElastiCache to reduce server load and speed-up popular requests.
The GWAS Explorer frontend website is built with React NodeJS. All user interface components are developed with Bootstrap. Plots for visualization of participant descriptive characteristics and association data are built in Plotly.js as well as custom solutions; for example, Manhattan plots and gene tracks are built in custom canvas. The quantile-quantile (Q-q), principal component (PC) plots and frequency plots are generated using Plotly.js and the bubble charts in the Browse Phenotypes section are produced using custom D3.js. The Apache service handles and serves all incoming web requests.
[ad_2]
Source link